About me
About me
A list of all the posts and pages found on the site. For you robots out there is an XML version available for digesting as well.
About me
Published:
Version 1.0 of the GGPONC medical text corpus is now available to researchers: Link
Published:
Our paper describing version 1.0 of the GGPONC corpus will be presented at the LOUHI workshop at EMNLP.
Published:
We have published an article in the German Digital Health magazine “Gesundhyte.de” about the role of NLP in evidence-based medicine and our GGPONC corpus.
Published:
Our paper “Knowledge bases and software support for variant interpretation in precision oncology” has been published in Briefings in Bioinformatics! Thanks to all collaborators from the HiGHmed consortium.
Published:
Our paper “Controversial Trials First: Identifying Disagreement Between Clinical Guidelines and New Evidence” featuring the Next Generation Evidence Browser will be presented at the AMIA Annual Symposium in November ‘21.
Published:
I am excited that our paper “Controversial Trials First: Identifying Disagreement Between Clinical Guidelines and New Evidence”, presented at the AMIA Annual Symposium received a Distinguished Paper Award.
Published:
I have organized a workshop with experts from both the German clinical NLP and the clinical guideline communities, to deepen the dialogue that we have started with our GGPONC project. Check out the event website for details.
Published:
Our paper for the new release of GGPONC 2.0 has been accepted at LREC! The new dataset is currently the largest, freely distributable annotated corpus of German medical text (1.87M tokens, 250K annotations). We also created baseline NER models with HuggingFace transformers.
Published:
Our contribution to the DisTEMIST shared task reached the 1st place in subtrack 2 (entity linking) and 2nd place in subtrack 1 (entity recognition). These are the results.
Published:
I gave a talk at the GIN 2022 Conference in Toronto about “Continuous Surveillance of Clinical Practice Guidelines through Natural Language Processing - Experiences from the GGPONC Project”. It was a very inspiring event with great conversations about the role NLP can play in guideline development and evidence synthesis.
Published:
We have published an implementation of FairEval on HuggingFace, which allows to get a detailed breakdown of errors for sequence labeling tasks (we consider mostly NER).
Published:
I am happy to announce that two papers got accepted at the International Conference of Artificial Intelligence in Medicine AIME 2023:
Published:
We have released xMEN - a Toolkit for Cross-Lingual Medical Entity Normalization on GitHub.
Prediction of defects in continuous steel casting
Predictive Business Process Monitoring
Development of interactive XR applications for HoloLens and other HMDs / mobile devices
Integrating Clinical Evidence with NLP
Cross-lingual Medical Entity Normalization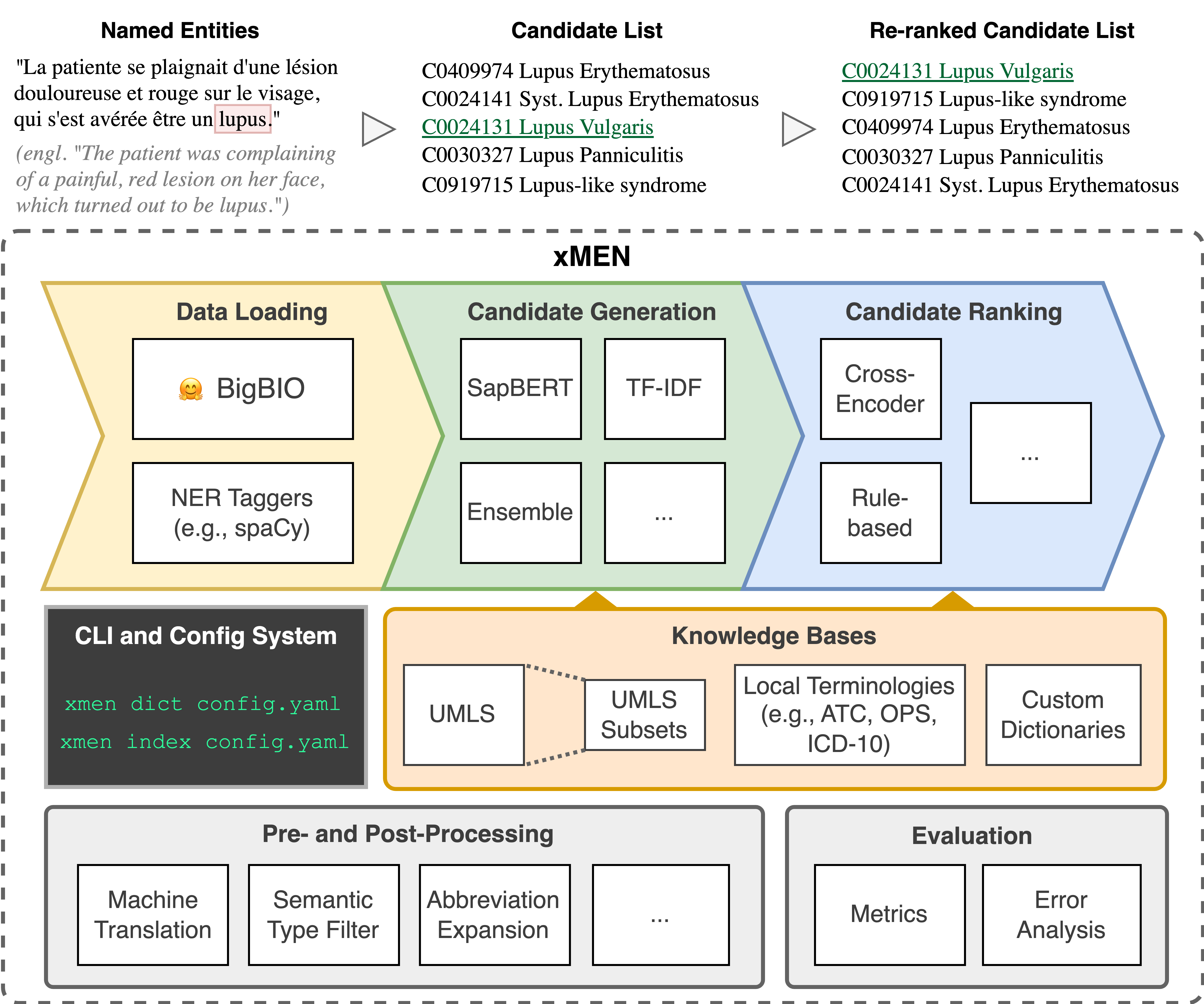
Clinical NLP for German
Published in LOUHI@EMNLP, 2020
Recommended citation: Florian Borchert*, Christina Lohr*, Luise Modersohn*, Thomas Langer, Markus Follmann, Jan Philipp Sachs, Udo Hahn, Matthieu-P. Schapranow. GGPONC: A Corpus of German Medical Text with Rich Metadata Based on Clinical Practice Guidelines. In Proceedings of the 11th International Workshop on Health Text Mining and Information Analysis, pp. 38–48. Online: Association for Computational Linguistics, 2020. (* equal contribution) [Data Access] [Code]
Published in Briefings in Bioinformatics, 2021
Recommended citation: Florian Borchert*, Andreas Mock*, Aurelie Tomczak*, Jonas Hügel, Samer Alkarkoukly, Alexander Knurr, Anna-Lena Volckmar, Albrecht Stenzinger, Peter Schirmacher, Jürgen Debus, Dirk Jäger, Thomas Longerich, Stefan Fröhling, Roland Eils, Nina Bougatf, Ulrich Sax, Matthieu-P Schapranow. Knowledge Bases and Software Support for Variant Interpretation in Precision Oncology, Briefings in Bioinformatics, Volume 22, Issue 6, November 2021, bbab134 (* equal contribution) IF = 11.6 https://doi.org/10.1093/bib/bbab134.
Published in 1st Conference on ICT for Health, Accessibility and Wellbeing, 2021
Recommended citation: Richard Henkenjohann, Benjamin Bergner, Florian Borchert, Nina Bougatf, Hauke Hund, Roland Eils, and Matthieu-P. Schapranow. An Engineering Approach towards Multi-Site Virtual Molecular Tumor Board Software Support. Proceedings of the 1st Conference on ICT for Health, Accessibility and Wellbeing. Springer International Publishing, 2021
Published in IEEE International Conference on Bioinformatics and Biomedicine, 2021
Recommended citation: Aadil Rasheed, Florian Borchert, Lasse Kohlmeyer, Richard Henkenjohann, and Matthieu-P. Schapranow. A Comparison of Concept Embeddings for German Clinical Corpora. IEEE International Conference on Bioinformatics and Biomedicine (BIBM), pp. 2314-2321, Online, 2021
Published in AMIA 2021 Annual Symposium, 2021
Recommended citation: Florian Borchert, Laura Meister, Thomas Langer, Markus Follmann, Bert Arnrich, and Matthieu-P. Schapranow. Controversial Trials First: Identifying Disagreement Between Clinical Guidelines and New Evidence, Proceedings of the AMIA Annual Symposium, pp. 237-246, San Diego, USA (2021) 🏆 Distinguished Paper Award [Link]
Published in LREC, 2022
Recommended citation: Florian Borchert, Christina Lohr, Luise Modersohn, Jonas Witt, Thomas Langer, Markus Follmann, Matthias Gietzelt, Bert Arnrich, Udo Hahn and Matthieu-P. Schapranow. GGPONC 2.0 - The German Clinical Guideline Corpus for Oncology: Curation Workflow, Annotation Policy, Baseline NER Taggers. LREC 2022 — Proceedings of the Language Resources and Evaluation Conference, pp. 3650‑3660. Marseille, France, European Language Resources Association, 2022 [Data Access] [Code]
Published in CLEF, 2022
Recommended citation: Florian Borchert and Matthieu-P. Schapranow. HPI-DHC @ BioASQ DisTEMIST: Spanish Biomedical Entity Linking with Pre-trained Transformers and Cross-lingual Candidate Retrieval. Proceedings of the Working Notes of CLEF 2022 - Conference and Labs of the Evaluation Forum, pp. 244-258. Bologna, Italy. 🏆 1st place DisTEMIST shared task (entity linking subtrack) [Link] [Code]
Published in AIME, 2023
Recommended citation: Sandro Steinwand*, Florian Borchert*, Silvia Winkler and Matthieu-P. Schapranow. GGTWEAK: Gene Tagging with Weak Supervision for German Clinical Text. In: Juarez, J.M., Marcos, M., Stiglic, G., Tucker, A. (eds) Artificial Intelligence in Medicine. AIME 2023. Lecture Notes in Computer Science, vol 13897. Springer, Cham [Code]
Published in AIME, 2023
Recommended citation: Julian Hugo, Susanne Ibing, Florian Borchert, Jan Philipp Sachs, Judy Cho, Ryan C. Ungaro and Erwin P. Böttinger. Machine Learning Based Prediction of Incident Cases of Crohn’s Disease Using Electronic Health Records from a Large Integrated Health System. In: Juarez, J.M., Marcos, M., Stiglic, G., Tucker, A. (eds) Artificial Intelligence in Medicine. AIME 2023. Lecture Notes in Computer Science, vol 13897. Springer, Cham 🏆 Best Student Paper
Published in Nature Scientific Data, 2023
Recommended citation: Phillip Richter-Pechanski, Philipp Wiesenbach, Dominic M. Schwab, Christina Kiriakou, Mingyang He, Michael M. Allers, Anna S. Tiefenbacher, Nicola Kunz, Anna Martynova, Noemie Spiller, Julian Mierisch, Florian Borchert, Charlotte Schwind, Norbert Frey, Christoph Dieterich & Nicolas A. Geis. A distributable German clinical corpus containing cardiovascular clinical routine doctor’s letters. Scientific Data 10, 207 (2023) [Data Access] IF = 10.8
Published in Health Informatics Journal, 2023
Recommended citation: Nektarios Ladas, Florian Borchert, Stefan Franz, Alina Rehberg, Natalia Strauch, Kim Katrin Sommer, Michael Marschollek, Matthias Gietzelt Programming techniques for improving rule readability for rule-based information extraction natural language processing pipelines of unstructured and semi-structured medical texts. Health Informatics Journal; 29(2) (2023)
Published in SN Computer Science, 2023
Recommended citation: Matthieu-P. Schapranow, Florian Borchert, Nina Bougatf, Hauke Hund, and Roland Eils. Software-Tool Support for Collaborative, Virtual, Multi-Site Molecular Tumor Boards. SN Computer Science 4, 358, 2023
Published in ACL, 2023
Recommended citation: Niklas Kämmer*, Florian Borchert*, Silvia Winkler, Gerard de Melo, and Matthieu-P. Schapranow Resolving Elliptical Compounds in German Medical Text.. In: The 22nd Workshop on Biomedical Natural Language Processing and BioNLP Shared Tasks, pages 292–305, Toronto, Canada. Association for Computational Linguistics
Published in ACL, 2023
Recommended citation: Ignacio Llorca, Florian Borchert, Matthieu-P. Schapranow A Meta-dataset of German Medical Corpora: Harmonization of Annotations and Cross-corpus NER Evaluation. In: Proceedings of the 5th Clinical Natural Language Processing Workshop, pages 171–181, Toronto, Canada. Association for Computational Linguistics
Published in CLEF, 2023
Recommended citation: Florian Borchert, Ignacio Llorca, Matthieu-P. Schapranow Cross-Lingual Candidate Retrieval and Re-ranking for Biomedical Entity Linking. In: Experimental IR Meets Multilinguality, Multimodality, and Interaction. CLEF 2023. Lecture Notes in Computer Science, vol 14163. Springer, Cham 🏆 Best of Labs (BioASQ, CLEF 2022)
Published in medRxiv, 2023
Recommended citation: Linea Schmidt, Susanne Ibing, Florian Borchert, Julian Hugo, Allison Marshall, Jellyana Peraza, Judy H. Cho, Erwin P. Böttinger, Ryan C. Ungaro Extraction of Crohn’s Disease Clinical Phenotypes from Clinical Text Using Natural Language Processing. medRxiv 2023.10.16.23297099 (2023)
Published in arxiv, 2023
Recommended citation: Danielly de Paula, Florian Borchert, Ariane Sasso, Falk Uebernickel Understanding emotions in the context of IT-based self-monitoring . arXiv preprint arXiv:2311.05449 (2023).
Published in BioCreative, 2023
Recommended citation: Florian Borchert, Ignacio Llorca, Matthieu-P. Schapranow. HPI-DHC @ BC8 SympTEMIST Track: Detection and Normalization of Symptom Mentions with SpanMarker and xMEN. Proceedings of the BioCreative VIII Challenge and Workshop: Curation and Evaluation in the Era of Generative Models. New Orleans, USA (2023) 🏆 1st place SympTEMIST shared task (entity linking subtrack) [Code]
Published in NTCIR-17, 2023
Recommended citation: Smilla Fox, Martin Preiß, Florian Borchert, Aadil Rasheed, Matthieu-P. Schapranow HPIDHC at NTCIR-17 MedNLP-SC: Data Augmentation and Ensemble Learning for Multilingual Adverse Drug Event Detection. NTCIR 17 Conference: Proceedings of the 17th NTCIR Conference on Evaluation of Information Access Technologies. pp. 185–192 (2023)
Published in Elsevier, 2024
Recommended citation: Keno K. Bressem, Jens-Michalis Papaioannou, Paul Grundmann, Florian Borchert, Lisa C. Adams, Leonhard Liu, Felix Busch, Lina Xu, Jan P. Loyen, Stefan M. Niehues, Moritz Augustin, Lennart Grosser, Marcus R. Makowski, Hugo JWL. Aerts, Alexander Löser. medBERT.de: A Comprehensive German BERT Model for the Medical Domain. Expert Systems with Applications (2024): 121598 [Hugging Face Model] IF = 8.5
Published in Database, 2024
Recommended citation: Florian Borchert, Ignacio Llorca, Matthieu-P. Schapranow. Improving biomedical entity linking for complex entity mentions with LLM-based text simplification. Database, Volume 2024, 2024, baae067 [Code]
Published in Springer, 2024
Recommended citation: Nico Steckhan, Raphaela Ring, Florian Borchert, Daniela A. Koppold Triangulation of Questionnaires, Qualitative Data and Natural Language Processing: A Differential Approach to Religious Bahá’í Fasting in Germany. J Relig Health, 63, 3360–3373 (2024)
Published in medRxiv, 2024
Recommended citation: Florian Borchert, Paul Wullenweber, Annika Oeser, Nina Kreuzberger, Torsten Karge, Thomas Langer, Nicole Skoetz, Lothar H. Wieler, Matthieu-P. Schapranow, Bert Arnrich. Next Generation Evidence: High-Precision Information Retrieval for Rapid Clinical Guideline Updates. medRxiv 2024.12.02.24318184 (2024)
Published in AI in Medicine, 2025
Recommended citation: Susanne Ibing, Julian Hugo, Florian Borchert, Linea Schmidt, Caroline Benson, Allison Marshall, Colleen Chasteau, Ujunwa Korie, Diana Paguay, Jan Philipp Sachs, Bernhard Y. Renard, Judy H. Cho, Erwin P. Böttinger, Ryan C. Ungaro. Electronic Health Records-based identification of newly diagnosed Crohn’s Disease cases. Artificial Intelligence in Medicine, Volume 159, January 2025, 103032 IF = 6.1
Published in JAMIA Open, 2025
Recommended citation: Florian Borchert, Ignacio Llorca, Roland Roller, Bert Arnrich, Matthieu-P. Schapranow xMEN: A Modular Toolkit for Cross-Lingual Medical Entity Normalization. JAMIA Open, Volume 8, Issue 1, ooae147 (2025). [Code] [Hugging Face Models]